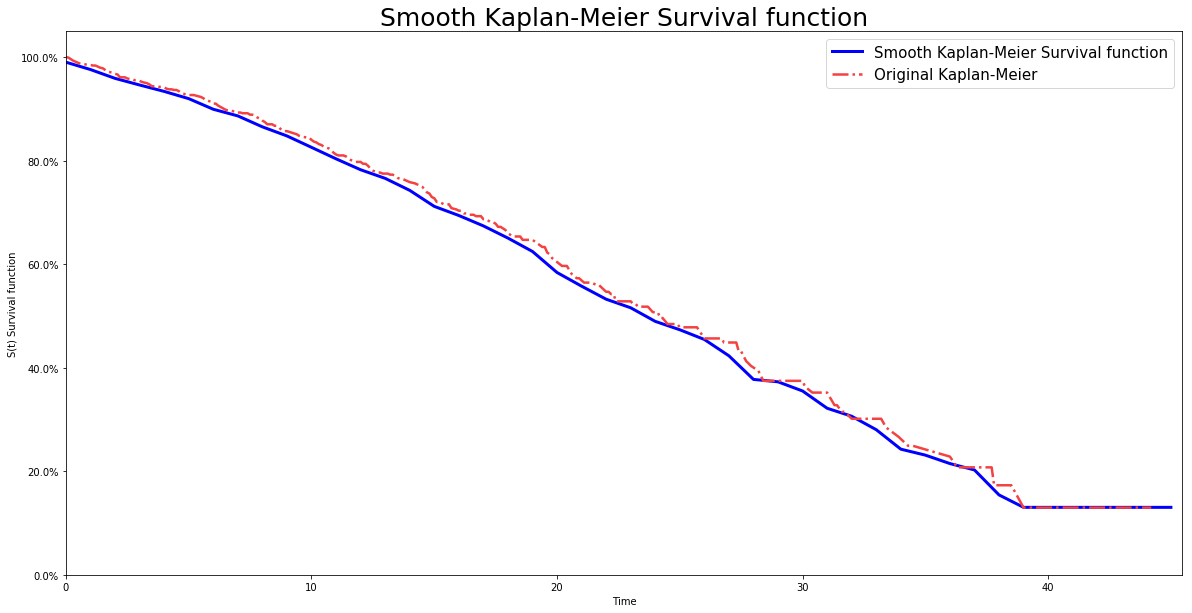
Smooth Kaplan Meier model
Smooth Kaplan Meier model is computed by using kernel smoothing to obtained a smooth estimator.
Instance
To create an instance, use pysurvival.models.non_parametric.SmoothKaplanMeierModel.
Attributes
cumulative_hazard: array-like -- representation of the cumulative hazard function of the modelhazard: array-like -- representation of the hazard function of the modelsurvival: array-like -- representation of the Survival function of the modeltimes: array-like -- representation of the time axis of the modeltime_buckets: array-like -- representation of the time axis of the model using time bins, which are represented by
Methods
__init__ - Initialize the estimator
SmoothKaplanMeierModel(bandwidth=0.1, kernel='normal')
Parameters:
-
bandwidth: double (default=0.1) -- controls the degree of the smoothing. The smaller it is the closer to the original KM the function will be, but it will increase the computation time. If it is very large, the resulting model will be smoother than the estimator of KM, but it will stop being as accurate. -
kernel: str (default='normal') -- defines the type of kernel the model will be using. Here are the possible options:Uniform: if elseEpanechnikov: if elseNormal:Biweight: if elseTriweight: if elseCosine: if else
fit - Fit the estimator based on the given parameters
fit(T, E, weights = None, alpha=0.05)
Parameters:
-
T: array-like -- target values describing the time when the event of interest or censoring occurred. -
E: array-like -- values that indicate if the event of interest occurred i.e.: E[i]=1 corresponds to an event, and E[i] = 0 means censoring, for all i. -
weights: array-like (default = None) -- array of weights that are assigned to individual samples. If not provided, then each sample is given a unit weight. -
alpha: float (default = 0.05) -- confidence level
Returns:
self: object
predict_density - Predicts the probability density function at a specified time t
predict_density(t)
Parameters:
t: double -- time at which the prediction should be performed.
Returns:
density: double -- prediction of the probability density function at t
predict_hazard - Predicts the hazard function at a specified time t
predict_hazard(t)
Parameters:
t: double -- time at which the prediction should be performed.
Returns:
hazard: double -- prediction of the hazard function at t
predict_survival - Predicts the survival function at a specified time t
predict_survival(t)
Parameters:
t: double -- time at which the prediction should be performed.
Returns:
survival: double -- prediction of the survival function at t
Example
# Importing modules import numpy as np from matplotlib import pyplot as plt from pysurvival.models.non_parametric import SmoothKaplanMeierModel from pysurvival.utils.display import display_non_parametric # %matplotlib inline #Uncomment when using Jupyter # Generating random times and event indicators T = np.round(np.abs(np.random.normal(10, 10, 1000)), 1) E = np.random.binomial(1, 0.3, 1000) # Initializing the SmoothKaplanMeierModel skm_model = SmoothKaplanMeierModel(bandwidth=1., kernel='Cosine') # Fitting the model and display the survival function and confidence intervals skm_model.fit(T, E, alpha=0.95) # Displaying the survival function and confidence intervals display_non_parametric(skm_model)