Simulation models
PySurvival can generate random survival times based on the most commonly used distributions such as:
- Exponential
- Weibull
- Gompertz
- Log-Logistic
- Lognormal
Instance
To create an instance, use pysurvival.models.simulations.SimulationModel.
Attributes
alpha: double -- the scale parameterbeta: double -- the shape parametercensored_parameter: double -- coefficient used to calculate the censored distribution.risk_type: string -- Defines the type of risk function.risk_parameter: double -- scaling coefficient of the risk scoresurvival_distribution: string -- Defines a known survival distribution.times: array-like -- representation of the time axis of the modeltime_buckets: array-like -- representation of the time axis of the model using time bins, which are represented by
API
__init__ - Initialization
SimulationModel( survival_distribution = 'exponential', risk_type = 'linear',
censored_parameter = 1., alpha = 1, beta = 1., bins = 100,
risk_parameter = 1.)
Parameters:
-
survival_distribution: string (default = 'exponential') -- Defines a known survival distribution. The available distributions are:ExponentialWeibullGompertzLog-LogisticLog-Normal
-
risk_type: string (default='linear') -- Defines the type of risk function: - Linear - Square - Gaussian -
censored_parameter: double (default = 1.) -- Coefficient used to calculate the censored distribution. This distribution is a normal such that N(loc=censored_parameter, scale=5) -
alpha: double (default = 1.) -- the scale parameter -
beta: double (default = 1.) -- the shape parameter -
bins: int (default=100) -- the number of bins of the time axis -
risk_parameter: double (default = 1.) -- Scaling coefficient for the risk score which can be written as follow:linear:square:gaussian:
generate_data - Generating a dataset of simulated survival times from a given distribution through the hazard function using the Cox model
generate_data(num_samples = 100, num_features = 3, feature_weights=None)
Parameters:
-
num_samples: int (default=100) -- Number of samples to generate -
num_features: int (default=3) -- Number of features to generate -
feature_weights: array-like (default=None) -- list of the coefficients of the underlying Cox-Model. The features linked to each coefficient are generated from random distribution from the following list:- binomial
- chisquare
- exponential
- gamma
- normal
- uniform
- laplace
If None then
feature_weights = [1.]*num_features
Returns:
dataset: pandas.DataFrame -- dataset of simulated survival times, event status and features
predict_hazard - Predicts the hazard function
predict_hazard(x, t = None)
Parameters:
-
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it -
t: double (default=None) -- time at which the prediction should be performed. If None, then it returns the function for all available t.
Returns:
hazard: numpy.ndarray -- array-like representing the prediction of the hazard function
predict_risk - Predicts the risk score
predict_risk(x)
Parameters:
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it
Returns:
risk_score: numpy.ndarray -- array-like representing the prediction of the risk score
predict_survival - Predicts the survival function
predict_survival(x, t = None)
Parameters:
-
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it -
t: double (default=None) -- time at which the prediction should be performed. If None, then return the function for all available t.
Returns:
survival: numpy.ndarray -- array-like representing the prediction of the survival function
Example
Let's now see how to generate a dataset designed for survival analysis.
import pandas as pd from pysurvival.models.simulations import SimulationModel %pylab inline # Initializing the simulation model sim = SimulationModel( survival_distribution = 'gompertz', risk_type = 'linear', censored_parameter = 5.0, alpha = 0.01, beta = 5., ) # Generating N Random samples N = 1000 dataset = sim.generate_data(num_samples = N, num_features=5) # Showing a few data-points dataset.head(2)
We can now see an overview of the data:
| x_1 | x_2 | x_3 | x_4 | x_5 | time | event |
|---|---|---|---|---|---|---|
| 3.956896 | 124.0 | 0.018274 | 57.480199 | -5.42258 | 0.024329 | 1.0 |
| 4.106100 | 117.0 | 0.111276 | 51.770875 | 4.105588 | 0.175530 | 1.0 |
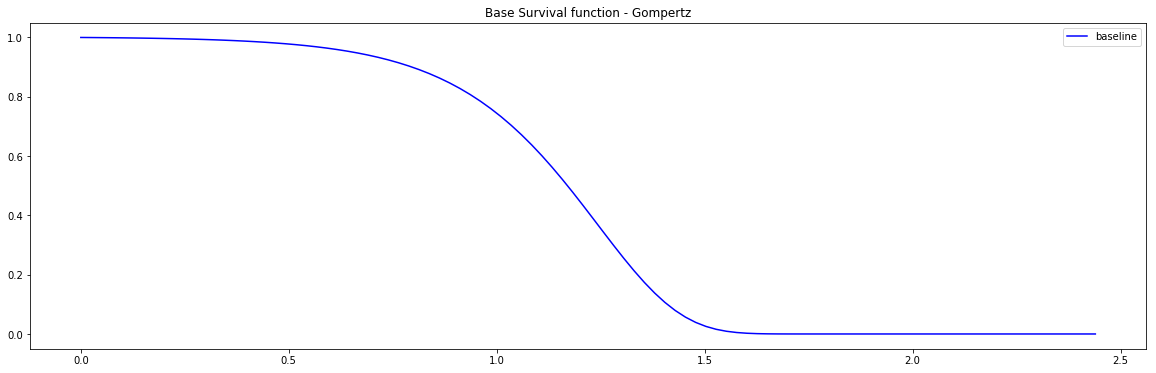
PySurvival also displays the Base Survival function of the Simulation model:
from pysurvival.utils.display import display_baseline_simulations display_baseline_simulations(sim, figure_size=(20, 6))