DeepSurv/Non-Linear model
The NonLinear CoxPH model was popularized by Katzman et al. in DeepSurv: Personalized Treatment Recommender System Using A Cox Proportional Hazards Deep Neural Network by allowing the use of Neural Networks within the original design and therefore introducing more modeling flexibility.
Instance
To create an instance, use pysurvival.models.semi_parametric.NonLinearCoxPHModel.
Attributes
baseline_hazard: array-like -- values of the hazard function whenbaseline_survival: array-like -- values of the survival function whenstructure: list of dictionaries -- Provides the structure of the MLP within the modeltimes: array-like -- representation of the time axis of the modeltime_buckets: array-like -- representation of the time axis of the model using time bins, which are represented by
Methods
__init__ - Initialization
NonLinearCoxPH(structure, auto_scaler=True)
Parameters:
-
structure: list of dictionaries -- Provides the structure of the MLP within the model.ex:
structure = [ {'activation': 'ReLU', 'num_units': 128}, {'activation': 'Tanh', 'num_units': 128}, ].Each dictionary corresponds to a fully connected hidden layer:
num_unitsis the number of hidden units in this layeractivationis the activation function that will be used. The list of all available activation functions can be found here.
In case there are more than one dictionary, each hidden layer will be applied in the resulting MLP, using the order it is provided in the structure .
-
auto_scaler: boolean (default=True) -- Determines whether a sklearn scaler should be automatically applied
fit - Fit the estimator based on the given parameters
fit(X, T, E, init_method = 'glorot_uniform', optimizer ='adam',
lr = 1e-4, num_epochs = 1000, dropout = 0.2, l2_smooth=1e-2,
batch_normalization=False, bn_and_dropout=False,
display_loss = True, verbose=True, extra_pct_time = 0.1)
Parameters:
-
X: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). -
T: array-like -- target values describing the time when the event of interest or censoring occurred. -
E: array-like -- values that indicate if the event of interest occurred i.e.: E[i]=1 corresponds to an event, and E[i] = 0 means censoring, for all i. -
init_method: str (default = 'glorot_uniform') -- Initialization method to use. Here are the possible options:glorot_uniform: Glorot/Xavier uniform initializerhe_uniform: He uniform variance scaling initializeruniform: Initializing tensors with uniform (-1, 1) distributionglorot_normal: Glorot normal initializer,he_normal: He normal initializer.normal: Initializing tensors with standard normal distributionones: Initializing tensors to 1zeros: Initializing tensors to 0orthogonal: Initializing tensors with a orthogonal matrix,
-
optimizer: str (default = 'adam') -- iterative method for optimizing a differentiable objective function. Here are the possible options:adadeltaadagradadamadamaxrmspropsparseadamsgd
-
lr: float (default=1e-4) -- learning rate used in the optimization -
num_epochs: int (default=1000) -- number of iterations in the optimization -
dropout: float (default=0.5) -- randomly setting a fraction rate of input units to 0 at each update during training time, which helps prevent overfitting. -
l2_reg: float (default=1e-4) -- L2 regularization parameter for the model coefficients -
batch_normalization: bool (default=True) -- whether or not applying Batch Normalization or not -
bn_and_dropout: bool (default=False) -- whether or not applying Batch Normalization and Dropout at the same time -
verbose: bool (default=True) -- whether or not producing detailed logging about the modeling
Returns:
- self : object
predict_hazard - Predicts the hazard function
predict_hazard(x, t = None)
Parameters:
-
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it -
t: double (default=None) -- time at which the prediction should be performed. If None, then it returns the function for all available t.
Returns:
hazard: numpy.ndarray -- array-like representing the prediction of the hazard function
predict_risk - Predicts the risk score
predict_risk(x)
Parameters:
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it
Returns:
risk_score: numpy.ndarray -- array-like representing the prediction of the risk score
predict_survival - Predicts the survival function
predict_survival(x, t = None)
Parameters:
-
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it -
t: double (default=None) -- time at which the prediction should be performed. If None, then return the function for all available t.
Returns:
survival: numpy.ndarray -- array-like representing the prediction of the survival function
Example
Let's now take a look at how to use the NonLinear CoxPH model on a simulation dataset generated from a parametric model.
#### 1 - Importing packages import numpy as np import pandas as pd from matplotlib import pyplot as plt from sklearn.model_selection import train_test_split from pysurvival.models.simulations import SimulationModel from pysurvival.models.semi_parametric import NonLinearCoxPHModel from pysurvival.utils.metrics import concordance_index from pysurvival.utils.display import integrated_brier_score #%pylab inline #### 2 - Generating the dataset from a nonlinear Weibull parametric model # Initializing the simulation model sim = SimulationModel( survival_distribution = 'weibull', risk_type = 'gaussian', censored_parameter = 2.1, alpha = 0.1, beta=3.2 ) # Generating N random samples N = 1000 dataset = sim.generate_data(num_samples = N, num_features=3) # Showing a few data-points dataset.head(2)
| x_1 | x_2 | x_3 | time | event |
|---|---|---|---|---|
| -13.851194 | 0.324245 | -0.153511 | 1.925692 | 1. |
| -0.094204 | 0.050048 | -0.214262 | 0.000000 | 0. |
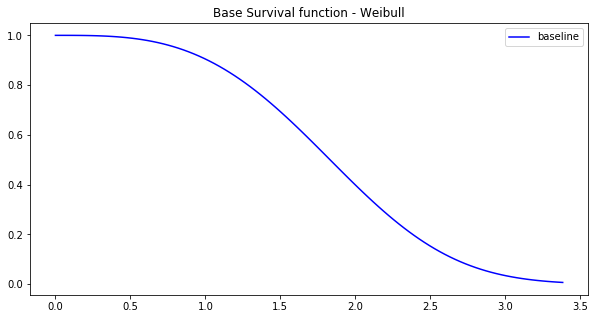
pySurvival also displays the Base Survival function of the Simulation model:
from pysurvival.utils.display import display_baseline_simulations display_baseline_simulations(sim, figure_size=(10, 5))
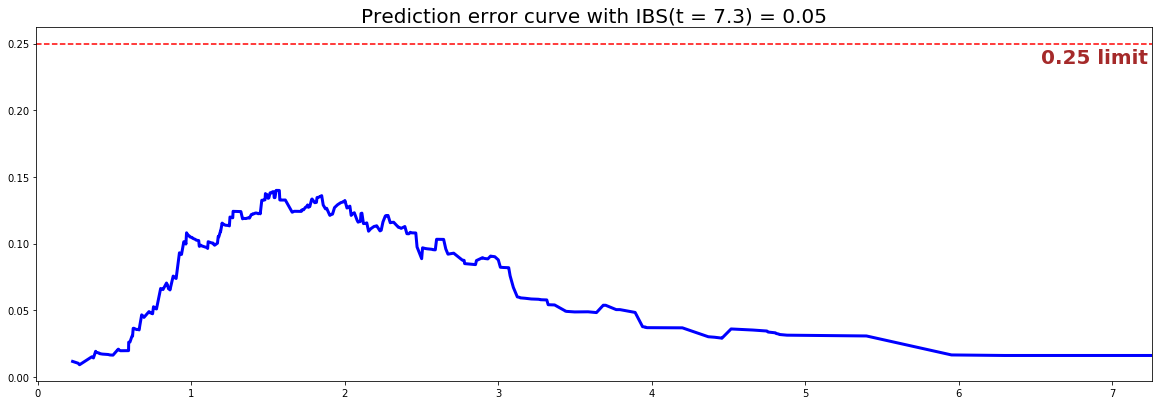
#### 3 - Creating the modeling dataset # Defining the features features = sim.features # Building training and testing sets # index_train, index_test = train_test_split( range(N), test_size = 0.2) data_train = dataset.loc[index_train].reset_index( drop = True ) data_test = dataset.loc[index_test].reset_index( drop = True ) # Creating the X, T and E input X_train, X_test = data_train[features], data_test[features] T_train, T_test = data_train['time'].values, data_test['time'].values E_train, E_test = data_train['event'].values, data_test['event'].values #### 4 - Creating an instance of the NonLinear CoxPH model and fitting the data. # Defining the MLP structure. Here we will build a 1-hidden layer # with 150 units and `BentIdentity` as its activation function structure = [ {'activation': 'BentIdentity', 'num_units': 150}, ] # Building the model nonlinear_coxph = NonLinearCoxPHModel(structure=structure) nonlinear_coxph.fit(X_train, T_train, E_train, lr=1e-3, init_method='xav_uniform') #### 5 - Cross Validation / Model Performances c_index = concordance_index(nonlinear_coxph, X_test, T_test, E_test) #0.81 print('C-index: {:.2f}'.format(c_index)) ibs = integrated_brier_score(nonlinear_coxph, X_test, T_test, E_test, t_max=10, figure_size=(20, 6.5) ) print('IBS: {:.2f}'.format(ibs))
We can see that the c-index is well above 0.5 and that the Prediction error curve is below the 0.25 limit, thus the model yields great performances. thus the model is likely to yield great performances.
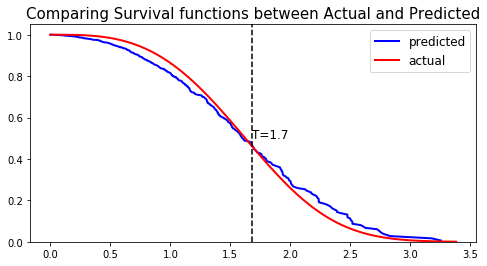
We can show this by randomly selecting datapoints and comparing the actual and predicted survival functions, computed by the simulation model and the Nonlinear CoxPH respectively.
#### 6 - Comparing actual and predictions # Initializing the figure fig, ax = plt.subplots(figsize=(8, 4)) # Randomly extracting a data-point that experienced an event choices = np.argwhere((E_test==1.)&(T_test>=1)).flatten() k = np.random.choice( choices, 1)[0] # Saving the time of event t = T_test[k] # Computing the Survival function for all times t predicted = nonlinear_coxph.predict_survival(X_test.values[k, :]).flatten() actual = sim.predict_survival(X_test.values[k, :]).flatten() # Displaying the functions plt.plot(nonlinear_coxph.times, predicted, color='blue', label='predicted', lw=2) plt.plot(sim.times, actual, color = 'red', label='actual', lw=2) # Actual time plt.axvline(x=t, color='black', ls ='--') ax.annotate('T={:.1f}'.format(t), xy=(t, 0.5), xytext=(t, 0.5), fontsize=12) # Show everything title = "Comparing Survival functions between Actual and Predicted" plt.legend(fontsize=12) plt.title(title, fontsize=15) plt.ylim(0, 1.05) plt.show()