Kernel SVM model
The Kernel SVM model available in PySurvival is an adaptation of the work of Sebastian Polsterl et al..
Instance
To create an instance, use pysurvival.models.svm.KernelSVMModel.
Methods
__init__ - Initialization
KernelSVMModel(kernel = "gaussian", scale=1., offset=0., degree=1.,
auto_scaler = True)
Parameters:
-
kernel: str (default="gaussian") -- The type of kernel used to fit the model. Here's the list of available kernels:- Polynomial
- Gaussian
- Exponential
- Tanh
- Sigmoid
- Rational Quadratic
- Inverse Multiquadratic
- Multiquadratic
-
scale: float (default=1) -- Scale parameter of the kernel function -
offset: float (default=0) -- Offset parameter of the kernel function -
degree: float (default=1) -- Degree parameter of the polynomial/kernel function
fit - Fit the estimator based on the given parameters
fit(X, T, E, with_bias = True, init_method='glorot_normal', lr = 1e-2,
max_iter = 100, l2_reg = 1e-4, tol = 1e-3, verbose = True)
Parameters:
-
X: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). -
T: array-like -- target values describing the time when the event of interest or censoring occurred. -
E: array-like -- values that indicate if the event of interest occurred i.e.: E[i]=1 corresponds to an event, and E[i] = 0 means censoring, for all i. -
with_bias: bool (default=True) -- Whether a bias should be added -
init_method: str (default = 'glorot_uniform') -- Initialization method to use. Here are the possible options:glorot_uniform: Glorot/Xavier uniform initializerhe_uniform: He uniform variance scaling initializeruniform: Initializing tensors with uniform (-1, 1) distributionglorot_normal: Glorot normal initializer,he_normal: He normal initializer.normal: Initializing tensors with standard normal distributionones: Initializing tensors to 1zeros: Initializing tensors to 0orthogonal: Initializing tensors with a orthogonal matrix,
-
lr: float (default=1e-4) -- learning rate used in the optimization -
max_iter: int (default=100) -- maximum number of iterations in the Newton optimization -
l2_reg: float (default=1e-4) -- L2 regularization parameter for the model coefficients -
alpha: float (default=0.95) -- confidence level -
tol: float (default=1e-3) -- tolerance for stopping criteria -
verbose: bool (default=True) -- whether or not producing detailed logging about the modeling
Returns:
- self : object
predict_risk - Predicts the risk score
predict_risk(x, use_log=True)
Parameters:
-
x: array-like -- input samples; where the rows correspond to an individual sample and the columns represent the features (shape=[n_samples, n_features]). x should not be standardized before, the model will take care of it -
use_log: bool (default=False) -- whether or not appliying the log function to the risk values
Returns:
risk_score: numpy.ndarray -- array-like representing the prediction of the risk score
Example
Let's now see how to use the KernelSVMModel models on a simulation dataset generated from a parametric model.
#### 1 - Importing packages import numpy as np import pandas as pd from matplotlib import pyplot as plt from pysurvival.models.svm import KernelSVMModel from pysurvival.models.simulations import SimulationModel from pysurvival.utils.metrics import concordance_index from sklearn.model_selection import train_test_split from scipy.stats.stats import pearsonr # %pylab inline # to use in jupyter notebooks #### 2 - Generating the dataset from the parametric model # Initializing the simulation model sim = SimulationModel( survival_distribution = 'Log-Logistic', risk_type = 'square', censored_parameter = 1.1, alpha = 1.5, beta = 4) # Generating N Random samples N = 1000 dataset = sim.generate_data(num_samples = N, num_features = 4) # Showing a few data-points dataset.head(2)
We can now see an overview of the data:
| x_1 | x_2 | x_3 | x_4 | time | event |
|---|---|---|---|---|---|
| 13.234733 | 10.0 | 12.0 | 11.0 | 0.264510 | 1.0 |
| 4.694893 | 14.0 | 11.0 | 7.0 | 0.000026 | 1.0 |
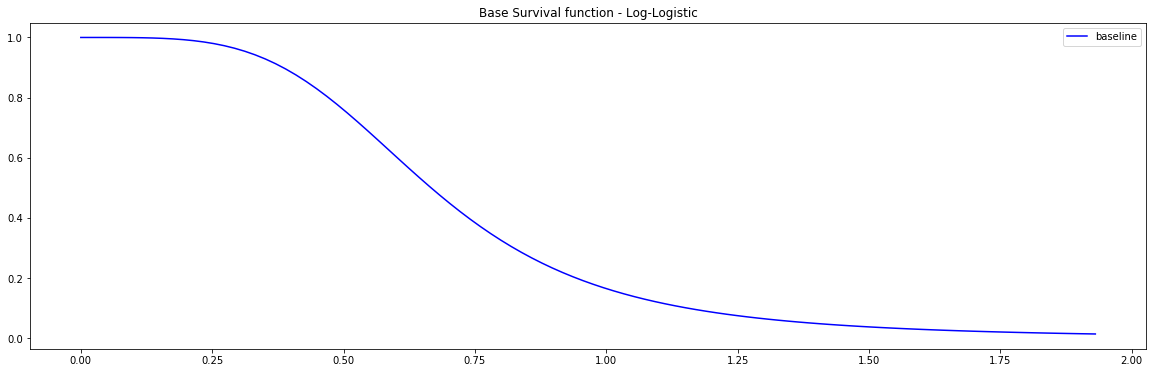
Pysurvival also displays the Base Survival function of the Simulation model:
from pysurvival.utils.display import display_baseline_simulations display_baseline_simulations(sim, figure_size=(20, 6))
#### 3 - Splitting the dataset into training and testing sets # Defining the features features = sim.features # Building training and testing sets # index_train, index_test = train_test_split( range(N), test_size = 0.2) data_train = dataset.loc[index_train].reset_index( drop = True ) data_test = dataset.loc[index_test].reset_index( drop = True ) # Creating the X, T and E input X_train, X_test = data_train[features], data_test[features] T_train, T_test = data_train['time'].values, data_test['time'].values E_train, E_test = data_train['event'].values, data_test['event'].values #### 4 - Creating an instance of the SurvivalSVM model and fitting the data. svm_model = KernelSVMModel(kernel='Gaussian', scale=0.25) svm_model.fit(X_train, T_train, E_train, init_method='orthogonal', with_bias = True, lr = 0.8, tol = 1e-3, l2_reg = 1e-4) #### 5 - Cross Validation / Model Performances c_index = concordance_index(svm_model, X_test, T_test, E_test) #0.89 print('C-index: {:.2f}'.format(c_index))
Because we cannot predict a survival function with KernelSVMModel, let's look at the
risk scores and see how correlated they are to the actual risk scores generated from the Simulation model.
#### 6 - Comparing the model predictions to Actual risk score # Comparing risk scores svm_risks = svm_model.predict_risk(X_test) actual_risks = np.log(sim.predict_risk(X_test).flatten()) print("corr={:.4f}, p_value={:.5f}".format(*pearsonr(svm_risks, actual_risks))) # corr=-0.7519, p_value=0.00000